× nearby
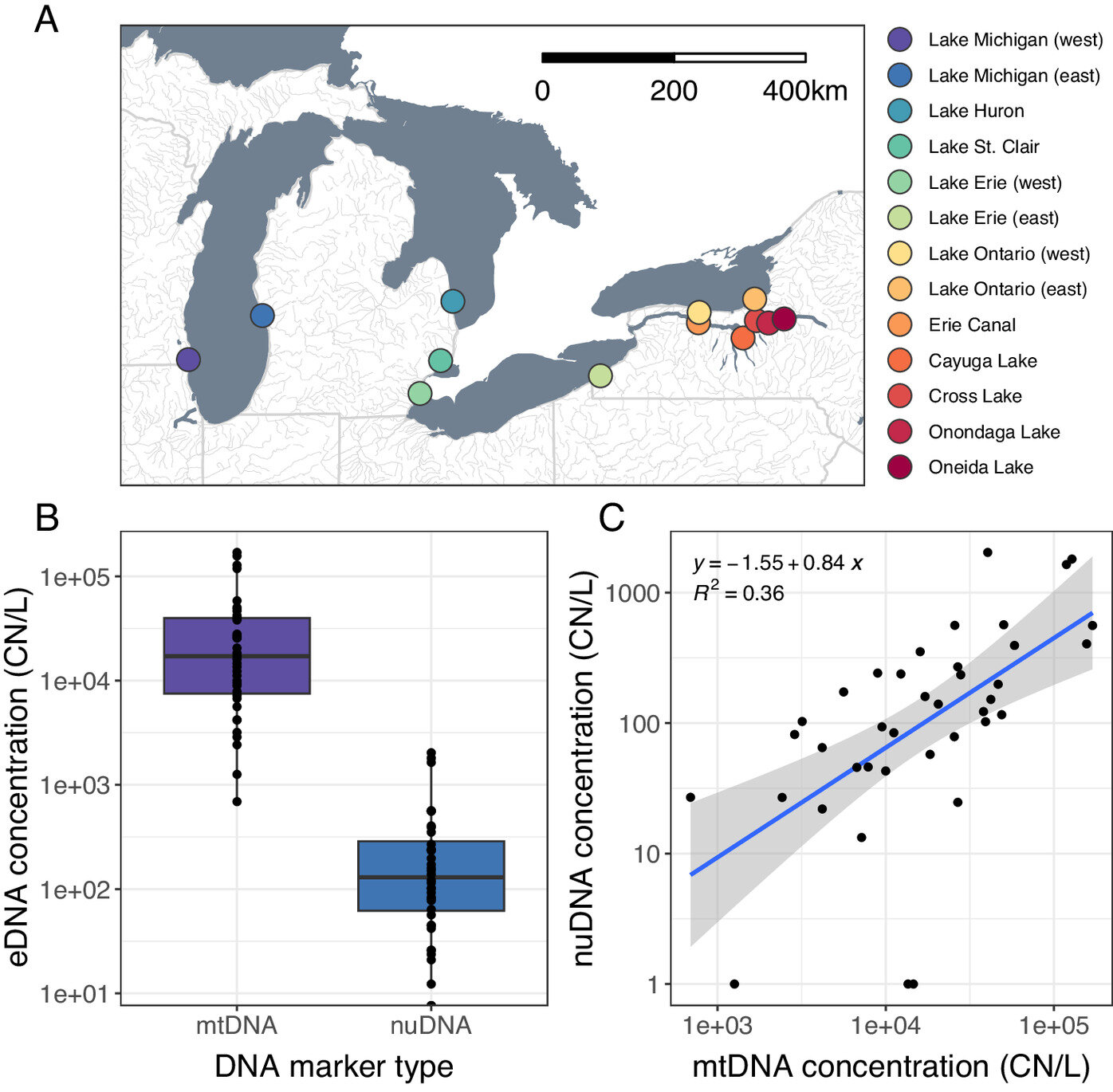
Results of a qPCR assay measuring mtDNA and nuDNA concentrations in eDNA samples. (A) Sample locations of Round Goby tissues and eDNA; (B) The concentration (copy number / L) of mtDNA (purple) in eDNA was significantly higher than the concentration of nuDNA (blue); (C) The log-translated mtDNA and nuDNA are strongly correlated with each other. Estimates (blue line) and 95% CI (shaded area) from a linear model for log-transformed DNA concentrations in each sample (C). Credit: Proceedings of the National Institute of Science (2023). DOI: 10.1073/pnas.2307345120
Ecologists have shown that the genetic material shed by different species in their environment can reveal not only the existence of a species but also a wide range of information about the genetics of all people.
Advances in environmental DNA (eDNA) are opening up new opportunities for the protection of endangered species and the control of invasive species.
“This breakthrough is part of an ongoing trend to learn more and more about eDNA, and this new study to detect genetic variation within a species,” said co-author David Lodge, Francis J. DiSalvo Director of the Cornell Atkinson Center for Sustainability. “To benefit biodiversity, we’re getting closer to what forensic scientists do every day at crime scenes.”
In a new study, published in Proceedings of the National Institute of ScienceThe researchers showed that their method was successful in field sampling of goby fish throughout the Great Lakes and the New York Finger Lakes.
The work builds on a pilot study in Cayuga Lake two years ago, when researchers took tissue samples from round goby fish and eDNA samples from the water the fish live in. They found that the two methods provided the same genetic information.
The first author of both studies is Kara Andres, Ph.D. ’22, a former graduate student in the Lodge lab and now a postdoctoral fellow at Washington University in St. Louis. He wrote the paper with co-authors Lodge, who is also a professor in the Department of Ecology and Evolutionary Biology in the College of Agriculture and Life Sciences, and Jose Andrés, a Cornell Atkinson faculty fellow and senior executive research fellow. the Cornell Environmental DNA and Genomics Core Facility.
In most animal cells, the nucleus contains two copies of the complete genetic code, but each cell contains 100 to 1,000 copies of a smaller, stripped-down version of the genetic code in the mitochondria. Most research on eDNA to date has focused on mitochondrial DNA, because it is likely to be the most abundant in environmental samples, Kara Andres said. While mitochondrial DNA is good at distinguishing between species, it provides less information than nuclear DNA about differences between species.
“The nuclear genome is very, very large and contains a lot of variation between species,” says Kara Andres. “When I started my Ph.D., we didn’t know anyone who tried to look for diversity in the nuclear genome in the context of eDNA sampling—we didn’t even know it was possible.”
In their Great Lakes study, which involved collecting water and tissue samples from goby fish in 13 locations from Lake Michigan to Oneida Lake, the researchers found that their eDNA sampling method could be used to detect nuclear genetic diversity, making it easier genetic analysis. differences and differences between species. This information is useful for natural resource managers because it can help them track the origins of new invasive species and prevent further invasions or reduce the risk by determining how invasive species travel and how to contain them.
This breakthrough could help scientists understand the status of endangered species without requiring the capture of already rare and vulnerable species. Species experiencing population declines can be affected by the loss of genetic diversity, and eDNA can allow researchers to predict those declines, Kara Andres said.
“It’s a big step in unlocking the full potential of genomics techniques when applied to marine eDNA samples,” says Jose Andrés. “In the near future, I expect that this technique will allow us to study the nature and health of rare species. I believe this has profound implications, especially in marine environments.”
More information:
Kara J. Andres et al, Environmental DNA reveals genetic diversity and population structure of invasive species in the Laurentian Great Lakes, Proceedings of the National Institute of Science (2023). DOI: 10.1073/pnas.2307345120
Journal information:
Proceedings of the National Institute of Science
#Environmental #DNA #sequencing #identifies #genetic #diversity #invasive #fish