× nearby
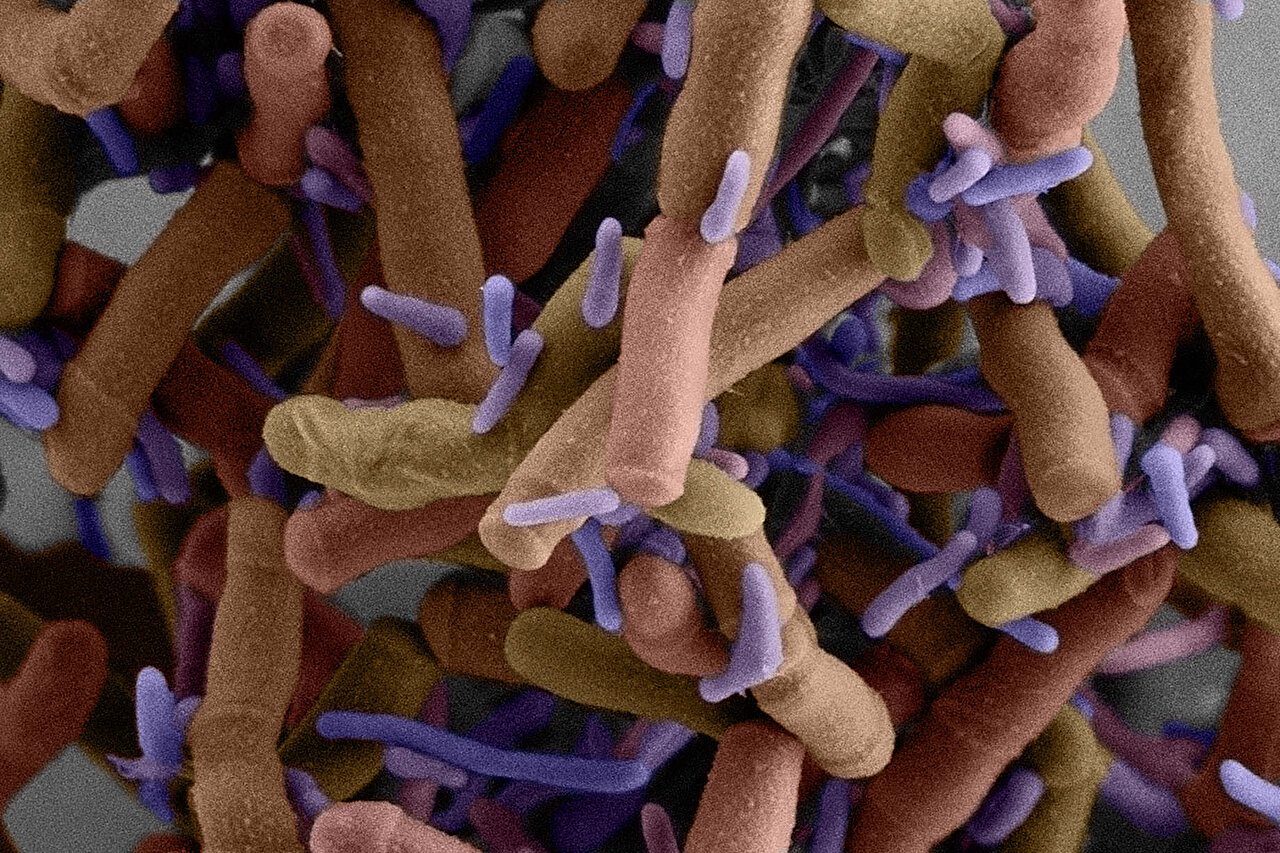
A scanning electron micrograph shows small purple cells of Patescibacteria growing on the surface of much larger cells. New research led by the lab of Joseph Mougous at UW Medicine in Seattle reveals their life cycle, their genes, and other molecular mechanisms that may be behind their unusual lifestyle. These epibiotic bacteria are Southlakia epibionticum. Credit: Yaxi Wang, Wai Pang Chan and Scott Braswell / University of Washington
Patescibacteria are a group of mysterious, tiny bacteria whose survival mechanisms are difficult to understand. Scientists can only cultivate a few species, but these bacteria are a diverse group found in many places.
A few species of Patescibacteria that researchers can grow in the lab live on the cell surface of another, larger microbe. Patescibacteria lack the genes needed to make many of the molecules necessary for life, such as the amino acids that make up proteins, the fatty acids that make up membranes, and the nucleotides in DNA. This has led researchers to think that many of them depend on other bacteria to grow.
In a study published in A cell, researchers present the first insight into the molecular mechanisms behind the unusual Patescibacteria lifestyle. This breakthrough was made possible by the discovery of a way to genetically control these bacteria, a breakthrough that has opened up a world of potential new research methods.
“Although metagenomics can tell us which microbes live on and inside our bodies, DNA sequencing alone does not give us insight into their beneficial or harmful functions, especially in organisms that have not been described before,” said Nitin S. Baliga of the Center for Systems Biology in Seattle, who has contributed several computational and analytical procedures to the study.
“The ability to genetically disrupt Patescibacteria opens up the possibility of using a powerful analytical lens system to rapidly characterize the unique biology of obligate epibionts,” he added, referring to organisms that must live on something else to survive.
The groups behind the study, led by the lab of Joseph Mougous in the Department of Microbiology at the University of Washington School of Medicine and the Howard Hughes Medical Institute, were interested in Patescibacteria for several reasons.
They are among the most poorly understood bacterial DNA sequences from large-scale genetic analysis of genomes found in species-rich microbial communities from environmental sources. This genetic material is called “microbial dark matter” because little is known about the functions it encodes.
Microbial black may contain information about biological methods and possible biotechnology applications, according to the A cell paper. It also holds clues to the molecular functions that support the microbial ecosystem, and the cell biology of the various microbial species collected in that system.
The Patescibacteria group analyzed in this recent study is Saccharibacteria. These live in a variety of soil and water environments but are best known for living in the human mouth. They have been part of the human oral microbiome since at least the Middle Stone Age and have been linked to human oral health.
In the human mouth, Saccharibacteria need the company of Actinobacteria, which act as their hosts. To better understand the mechanisms that Saccharibacteria use to communicate with their hosts, researchers used genetic manipulation to identify all the genes necessary for Saccharibacterium to grow.
“We are very excited to get this glimpse of the unusual genetic activity of these bacteria,” said Mougous, a professor of microbiology. “By focusing our future studies on these genes, we hope to unravel the mystery of how Saccharibacteria exploit the bacteria that arrest their growth.”
Potential host interactions revealed in the study include cell structures that may help Saccharibacteria adhere to host cells, and a unique secretion system that may be used to transport nutrients.
Another application of the authors’ work was the production of Saccharibacteria cells that express fluorescent proteins. With these cells, the researchers created microscopic fluorescent images of Saccharibacteria growing with their bacteria.
“Time-lapse imaging of Saccharibacteria-host cell cultures has revealed the remarkable complexity of the lives of these unusual bacteria,” said S. Brook Peterson, a senior scientist in the Mougous lab.
Researchers have reported that some Saccharibacteria act as mother cells by attaching to the host cell and multiplying by budding to produce smaller offspring. These young ones go on to look for new host cells. Some of the offspring, then, become mother cells, while others appear to communicate unproductively to the host.
The researchers think that further genetic studies will open the door to a broader understanding of the roles of what they describe as “the rich storage of dark matter contained in these organisms” and may reveal unsuspected biological mechanisms.
This interdisciplinary and collaborative study was promoted by the newly formed Microbial Interactions & Microbiome Center (acronym mim_c), directed by Mougous. The mission of mim_c is to lower the barriers to microbiome research studies and to advance collaboration by connecting like-minded researchers from all disciplines. Here, mim_c was the catalyst to join the Mougous lab with oral microbiome expert Jeffrey McClean in the Department of Periodontics, UW School of Dentistry.
More information:
Joseph D. Mougous et al, Genetic modification of Patescibacteria provides insight into dark matter issues and epibiotic lifestyles, A cell (2023). DOI: 10.1016/j.cell.2023.08.017. www.cell.com/cell/fulltext/S0092-8674(23)00906-6
Journal information:
A cell
#Genetic #tools #investigate #microbial #dark #matter